Introduction to AirVeg
AirVeg.RmdThe goal of AirVeg is to make it easy to import and manipulate data from air quality models and measurements. Validation of simulations with respect to observations can be performed with the provided statistical scores.
In this example, we will perform a simple validation of predicted (model) data with observed measurements of ground level O3 concentrations in the city of Bologna. Please note that the dataset cited in the following are not available inside the package due to the size of the files.
Import data
Firstly, we load the AirVeg library. We also load the
data.table package as it will be used to manipulate columns
and values of the dataset:
library(AirVeg)
library(data.table)data.table provides a high-performance version of base
R’s data.frame. If you are not familiar with it, please
check the documentation.
Let’s assume we have a dataset of model data in the NetCDF
file BO_c_2015_SVR_hourly_01.nc. We can check the content
of the archive with the AirVeg::nc_summary() function:
nc_summary("../devtemp/data/BO/FARM/BO_c_2015_SVR_hourly_01.nc")
#>
#> --- netCDF archive info ---
#> CF Convention : COARDS
#> First deadline : 01/01/2015 03:00:00 UTC
#> Last deadline : 01/02/2015 00:00:00 UTC
#> Deadline interval (s) : 3600
#> Number of deadlines : 742
#> Number of gridpoints (x, y, z): 50, 50, 0
#> Coord of SW corner (km, km) : 660.500 4905.500
#> Coord of NE corner (km, km) : 709.500 4954.500
#> Grid cell sizes (x, y) : 1.00 1.00
#> List of 2D variables ( 4) : c_O3 c_NO2 c_PM25 c_PM10We notice that the archive contains hourly values of four pollutants
for the single month of January 2015. If we want to read the
Ozone in the the c_O3 variable, we can use the
AirVeg::nc_get_variable() function, as:
farmData <- nc_get_variable(input = "../devtemp/data/BO/FARM/BO_c_2015_SVR_hourly_01.nc",
variable = "c_O3",
tz = "Etc/GMT-1")We can use the data.table notation to check for the
content of farmData:
farmData[]
#> time y x c_O3
#> <POSc> <num> <num> <num>
#> 1: 2015-01-01 03:00:00 4905.5 660.5 13.69436
#> 2: 2015-01-01 03:00:00 4905.5 661.5 15.36455
#> 3: 2015-01-01 03:00:00 4905.5 662.5 16.47361
#> 4: 2015-01-01 03:00:00 4905.5 663.5 16.27375
#> 5: 2015-01-01 03:00:00 4905.5 664.5 15.30639
#> ---
#> 1854996: 2015-02-01 00:00:00 4954.5 705.5 20.36149
#> 1854997: 2015-02-01 00:00:00 4954.5 706.5 21.08920
#> 1854998: 2015-02-01 00:00:00 4954.5 707.5 21.57969
#> 1854999: 2015-02-01 00:00:00 4954.5 708.5 21.88763
#> 1855000: 2015-02-01 00:00:00 4954.5 709.5 22.32390We see that the variable is composed of 4 columns: time
with date and time; x and y with grid
coordinates; c_O3 with the concentration values of O3. Each line of the table contains all
the information about the given deadline.
The grid coordinates are expressed in kilometres. As it will
be clear in the following, it is convenient to convert then to
metres. In order to do so, we again use the compact notation of
data.table and perform an in-place
transformation.
farmData[, `:=`(x = x * 1000, y = y * 1000)]
farmData[]
#> time y x c_O3
#> <POSc> <num> <num> <num>
#> 1: 2015-01-01 03:00:00 4905500 660500 13.69436
#> 2: 2015-01-01 03:00:00 4905500 661500 15.36455
#> 3: 2015-01-01 03:00:00 4905500 662500 16.47361
#> 4: 2015-01-01 03:00:00 4905500 663500 16.27375
#> 5: 2015-01-01 03:00:00 4905500 664500 15.30639
#> ---
#> 1854996: 2015-02-01 00:00:00 4954500 705500 20.36149
#> 1854997: 2015-02-01 00:00:00 4954500 706500 21.08920
#> 1854998: 2015-02-01 00:00:00 4954500 707500 21.57969
#> 1854999: 2015-02-01 00:00:00 4954500 708500 21.88763
#> 1855000: 2015-02-01 00:00:00 4954500 709500 22.32390To perform a very simple validation of the modeled data, we need a
corresponding observations dataset. In this case we can use
AirVeg::import_data_brace to read observations data from a
local file with a subset of the BRACE database:
obsO3 <- import_data_brace("../devtemp/data/BO/BRACE/O3_2015_BRACE/",
tz = "Etc/GMT-1")
obsO3[]
#> Key: <cod_brace>
#> cod_brace datetime obs x y nomestaz
#> <int> <POSc> <int> <num> <num> <char>
#> 1: 803708 2015-01-01 00:00:00 23 687271.6 4928255 GIARDINI_MARGHERITA
#> 2: 803708 2015-01-01 01:00:00 23 687271.6 4928255 GIARDINI_MARGHERITA
#> 3: 803708 2015-01-01 02:00:00 NA 687271.6 4928255 GIARDINI_MARGHERITA
#> 4: 803708 2015-01-01 03:00:00 23 687271.6 4928255 GIARDINI_MARGHERITA
#> 5: 803708 2015-01-01 04:00:00 29 687271.6 4928255 GIARDINI_MARGHERITA
#> ---
#> 17516: 803719 2015-12-31 19:00:00 6 681741.6 4929951 VIA_CHIARINI
#> 17517: 803719 2015-12-31 20:00:00 4 681741.6 4929951 VIA_CHIARINI
#> 17518: 803719 2015-12-31 21:00:00 3 681741.6 4929951 VIA_CHIARINI
#> 17519: 803719 2015-12-31 22:00:00 4 681741.6 4929951 VIA_CHIARINI
#> 17520: 803719 2015-12-31 23:00:00 3 681741.6 4929951 VIA_CHIARINI
#> regione tipo zona quota lat lon cod_airbase
#> <char> <char> <char> <int> <num> <num> <char>
#> 1: Emilia-Romagna background urban 43 44.48333 11.35500 IT0892A
#> 2: Emilia-Romagna background urban 43 44.48333 11.35500 IT0892A
#> 3: Emilia-Romagna background urban 43 44.48333 11.35500 IT0892A
#> 4: Emilia-Romagna background urban 43 44.48333 11.35500 IT0892A
#> 5: Emilia-Romagna background urban 43 44.48333 11.35500 IT0892A
#> ---
#> 17516: Emilia-Romagna background suburban 56 44.50000 11.28611 IT2075A
#> 17517: Emilia-Romagna background suburban 56 44.50000 11.28611 IT2075A
#> 17518: Emilia-Romagna background suburban 56 44.50000 11.28611 IT2075A
#> 17519: Emilia-Romagna background suburban 56 44.50000 11.28611 IT2075A
#> 17520: Emilia-Romagna background suburban 56 44.50000 11.28611 IT2075AThe resulting output is again a data.table object which
contains measurements for two stations, as it can be easily
verified:
uniqueN(obsO3[, .(cod_brace)])
#> [1] 2Namely the stations are “Giardini Margherita” and “Via Chiarini” respectively of background/urban and background/suburban type:
unique(obsO3[, .(nomestaz, tipo, zona), by = .(cod_brace)])
#> Key: <cod_brace>
#> cod_brace nomestaz tipo zona
#> <int> <char> <char> <char>
#> 1: 803708 GIARDINI_MARGHERITA background urban
#> 2: 803719 VIA_CHIARINI background suburbanModel data extraction
We now need to extract the model data at the location of the two
measurement stations. This task can be accomplished with the help the
AirVeg::nc_extract() function. Here we need to specify the
predicted and observed dataset and the names of the relevant
columns.
farmDataBrace <- nc_extract(mydata = farmData,
name.x = "x", name.y = "y", name.time = "time", name.value = "c_O3",
stations = obsO3,
st.x = "x", st.y = "y", st.time = "datetime",
st.value = "obs", st.code = "cod_brace")In the first group of arguments, information about the model data are provided. The corresponding information for the observations are given in the second group of arguments.
In the example, it is used the default extraction method
(method="simple") i.e. the values of cell in which a point
falls are returned.
Alternatively a bilinear interpolation of the four nearest neighbours
can be performed by choosing the "bilinear" method.
farmDataBrace is a new data.table object
obtained merging the two orignal tables with both the observed data and
the model values extracted at the stations coordinates:
colnames(farmDataBrace)
#> [1] "cod_brace" "time" "x" "y" "c_O3"
#> [6] "obs" "nomestaz" "regione" "tipo" "zona"
#> [11] "quota" "lat" "lon" "cod_airbase"To improve readability, we can drop the unnecessary columns:
farmDataBrace[, `:=`(cod_airbase = NULL, lat = NULL, lon = NULL, quota = NULL,
regione = NULL, tipo = NULL, zona = NULL)]
farmDataBrace[]
#> Key: <cod_brace, time, x, y>
#> cod_brace time x y c_O3 obs
#> <int> <POSc> <num> <num> <num> <int>
#> 1: 803708 2015-01-01 03:00:00 687271.6 4928255 1.234904e+01 23
#> 2: 803708 2015-01-01 04:00:00 687271.6 4928255 2.263929e+01 29
#> 3: 803708 2015-01-01 05:00:00 687271.6 4928255 2.417113e+01 26
#> 4: 803708 2015-01-01 06:00:00 687271.6 4928255 1.723468e+01 21
#> 5: 803708 2015-01-01 07:00:00 687271.6 4928255 2.061162e+00 30
#> ---
#> 1480: 803719 2015-01-31 20:00:00 681741.6 4929951 2.941550e+00 4
#> 1481: 803719 2015-01-31 21:00:00 681741.6 4929951 3.731866e-01 5
#> 1482: 803719 2015-01-31 22:00:00 681741.6 4929951 2.186908e-04 5
#> 1483: 803719 2015-01-31 23:00:00 681741.6 4929951 2.199765e-02 5
#> 1484: 803719 2015-02-01 00:00:00 681741.6 4929951 2.679052e-01 5
#> nomestaz
#> <char>
#> 1: GIARDINI_MARGHERITA
#> 2: GIARDINI_MARGHERITA
#> 3: GIARDINI_MARGHERITA
#> 4: GIARDINI_MARGHERITA
#> 5: GIARDINI_MARGHERITA
#> ---
#> 1480: VIA_CHIARINI
#> 1481: VIA_CHIARINI
#> 1482: VIA_CHIARINI
#> 1483: VIA_CHIARINI
#> 1484: VIA_CHIARINIThe dataset now includes all the necessary columns to perform some
data validation. Specifically, the observed measurements are in the
obs column while the predicted data from FARM model are in
the c_O3 column.
Data validation
Now we can compute some validation scores, grouping the data by
station, using the compact data.table syntax:
farmDataBrace[, .(Ndata = countComplete(obs, c_O3),
bias = bias(obs = obs, pred = c_O3, use = "complete.obs"),
rmse = rmse(obs = obs, pred = c_O3, use = "complete.obs"),
corr = corr(obs = obs, pred = c_O3, use = "na.or.complete"),
nmse = nmse(obs = obs, pred = c_O3, use = "complete.obs"),
fb = fb(obs = obs, pred = c_O3, use = "complete.obs"),
fac2 = fac2(obs = obs, pred = c_O3, use = "complete.obs"),
ioa = ioa(obs = obs, pred = c_O3, use = "complete.obs")),
by = .(cod_brace)]
#> Key: <cod_brace>
#> cod_brace Ndata bias rmse corr nmse fb fac2
#> <int> <int> <num> <num> <num> <num> <num> <num>
#> 1: 803708 656 -2.788975 17.27184 0.4919275 1.820024 -0.2165626 0.2743902
#> 2: 803719 711 -2.350166 12.46522 0.5916661 1.034304 -0.1908693 0.3248945
#> ioa
#> <num>
#> 1: 0.6922993
#> 2: 0.7598378In many of the used statistical functions, we need to set the
"complete.obs" option to select only the rows where both
observed and predicted data are present. Otherwise, if some value is
missing (Not Available, NA), the result would be null:
farmDataBrace[, .(Ndata = countComplete(obs, c_O3),
bias = bias(obs = obs, pred = c_O3),
rmse = rmse(obs = obs, pred = c_O3),
corr = corr(obs = obs, pred = c_O3),
nmse = nmse(obs = obs, pred = c_O3),
fb = fb(obs = obs, pred = c_O3),
fac2 = fac2(obs = obs, pred = c_O3),
ioa = ioa(obs = obs, pred = c_O3)),
by = .(cod_brace)]
#> Key: <cod_brace>
#> cod_brace Ndata bias rmse corr nmse fb fac2 ioa
#> <int> <int> <num> <num> <num> <num> <num> <num> <num>
#> 1: 803708 656 NA NA NA NA NA NA NA
#> 2: 803719 711 NA NA NA NA NA NA NAData visualization
Eventually we can use the AirVeg::scatterPlot() graphic
function to visualize the observed and predicted (model) data we just
validated:
library(ggplot2)
scatterPlot(data = farmDataBrace, obs = "obs", mod = "c_O3") +
scale_x_continuous(limits = c(0, 100),
name = expression("Observated data ["~mu~g/m^3~"]")) +
scale_y_continuous(limits = c(0, 100), name = expression("Model data ["~mu~g/m^3~"]"))
#> Scale for x is already present.
#> Adding another scale for x, which will replace the existing scale.
#> Scale for y is already present.
#> Adding another scale for y, which will replace the existing scale.
#> Warning: Removed 117 rows containing missing values or values outside the scale range
#> (`geom_point()`).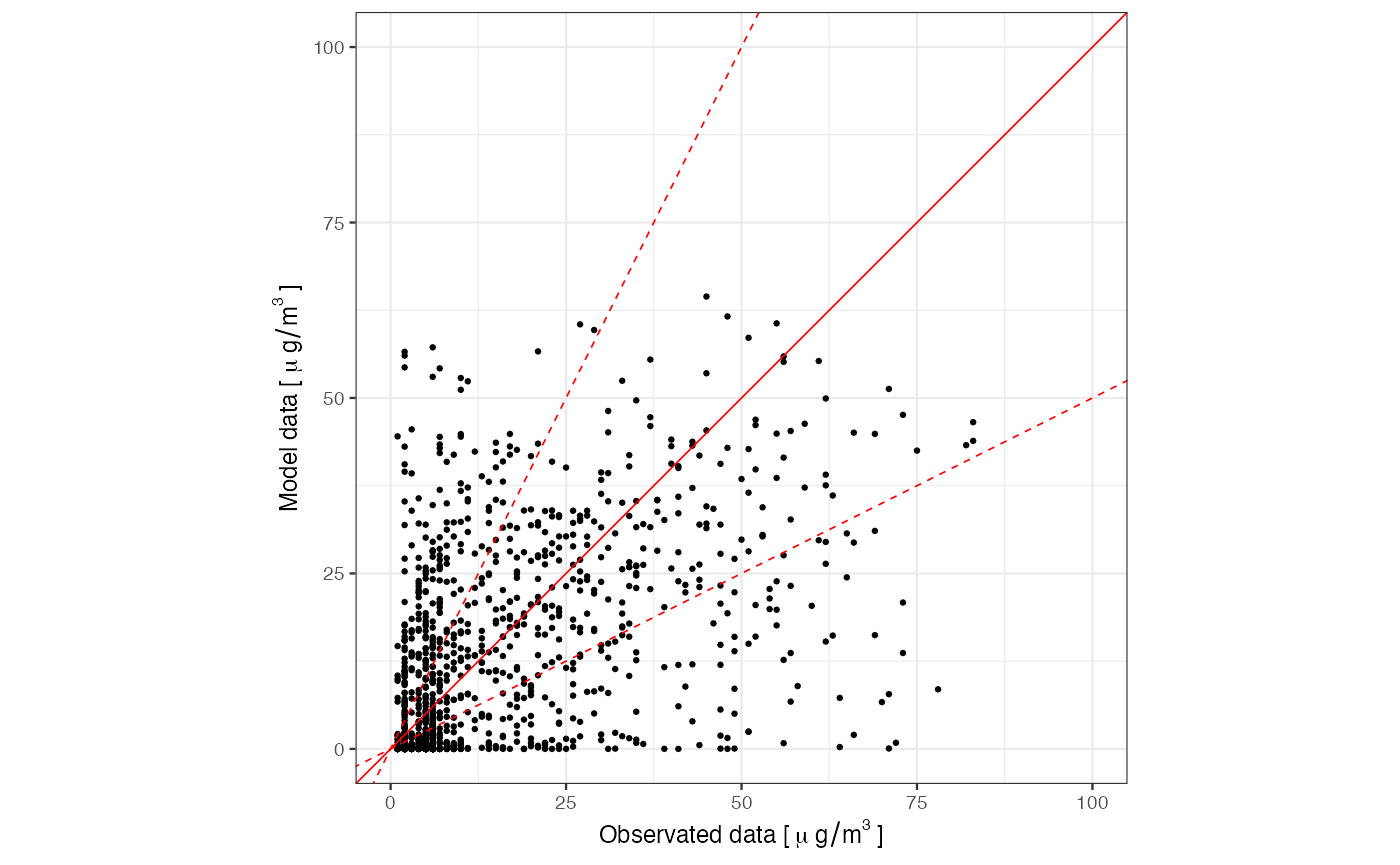
Please note that scatterPlot returns an object of class
ggplot, therefore it can be further customized by the
directives of the ggplot2 library, as with
scale_x_continuous and scale_y_continuous. In
order to do so, we had to explicitly load the library before running the
command. This is the case for all the graphics functions provided in
AirVeg.